WHAT IS LATIN-SEQ:
Latin-SEQ is a research project with the primary aim of providing genetic diagnosis to individuals affected by hereditary neuromuscular diseases (NMD) residing in Latin American (LATAM) countries. To achieve this, we utilise a range of genetic techniques, primarily «Next Generation Sequencing» (NGS), but also traditional sequencing using Sanger and MLPA techniques. This is an international and multicentre study involving over 60 hospitals across 18 countries, coordinated by the John Walton Muscular Dystrophy Research Centre (JWMDRC) at Newcastle University, United Kingdom.
Our commitment is to accelerate the genetic diagnosis process for these patients and expand knowledge about the causes and prevalence of neuromuscular diseases in different LATAM populations. Obtaining an early diagnosis offers patients numerous benefits: access to a follow-up plan and personalised therapy, clear expectations about the progression of their disease, the possibility of receiving genetic counselling for family members, the opportunity to be included in patient registries or associations, and participation in clinical trials if available.
Latin-SEQ promotes the creation of a robust network of hospital units and clinical centres with expertise in neuromuscular diseases, catering to both adult and paediatric patients across LATAM.
Latin-SEQ also includes virtual spaces for specialised training, joint discussion of clinical cases, and collaboration on research projects, bringing together neurologists, paediatric neurologists, neurophysiologists, geneticists, physiotherapists, and other healthcare professionals involved in the diagnosis and follow-up of patients with neuromuscular diseases.
OBJETIVES:
WHAT IS EXOME SEQUENCING:
Sequencing is a molecular technique that allows us to analyse a person’s DNA to “read” their genetic code. Previously, methods such as Sanger sequencing were used, which, although accurate, were slower as they only allowed for the sequencing of a single region of DNA (gene) at a time.
Nowadays, we use more advanced techniques like Next-Generation Sequencing (NGS). This new technology is faster and allows for the simultaneous sequencing of large regions of DNA in a short period of time, facilitating and accelerating genetic diagnosis. In the Latin-SEQ project, we use whole exome sequencing to obtain information from all coding regions, which are those that contain the instructions to produce all proteins. By comparing the exomes of an affected individual with a reference sequence, we can identify genetic variants that may be the cause of these neuromuscular diseases. Thanks to this technology, we can offer a faster and more accurate diagnosis, thus improving the care and quality of life of patients.
INCLUSION AND EXCLUSION CRITERIA:
All patients with a suspected genetic neuromuscular disease, even without a diagnosis, can be included in the Latin-SEQ project. There are no restrictions based on sex or age (both adult and paediatric patients are eligible), nor on the type of neuromuscular disease they present (affecting muscle, neuromuscular junction, or nerve). Only acquired diseases or those that cannot be diagnosed through exome analysis (such as DM1/2 and FSHD) will be excluded. The study will prioritise the analysis of families with multiple affected members and patients from ethnic minorities.
RECRUITMENT, SAMPLE COLLECTION, AND PHENOTYPING:
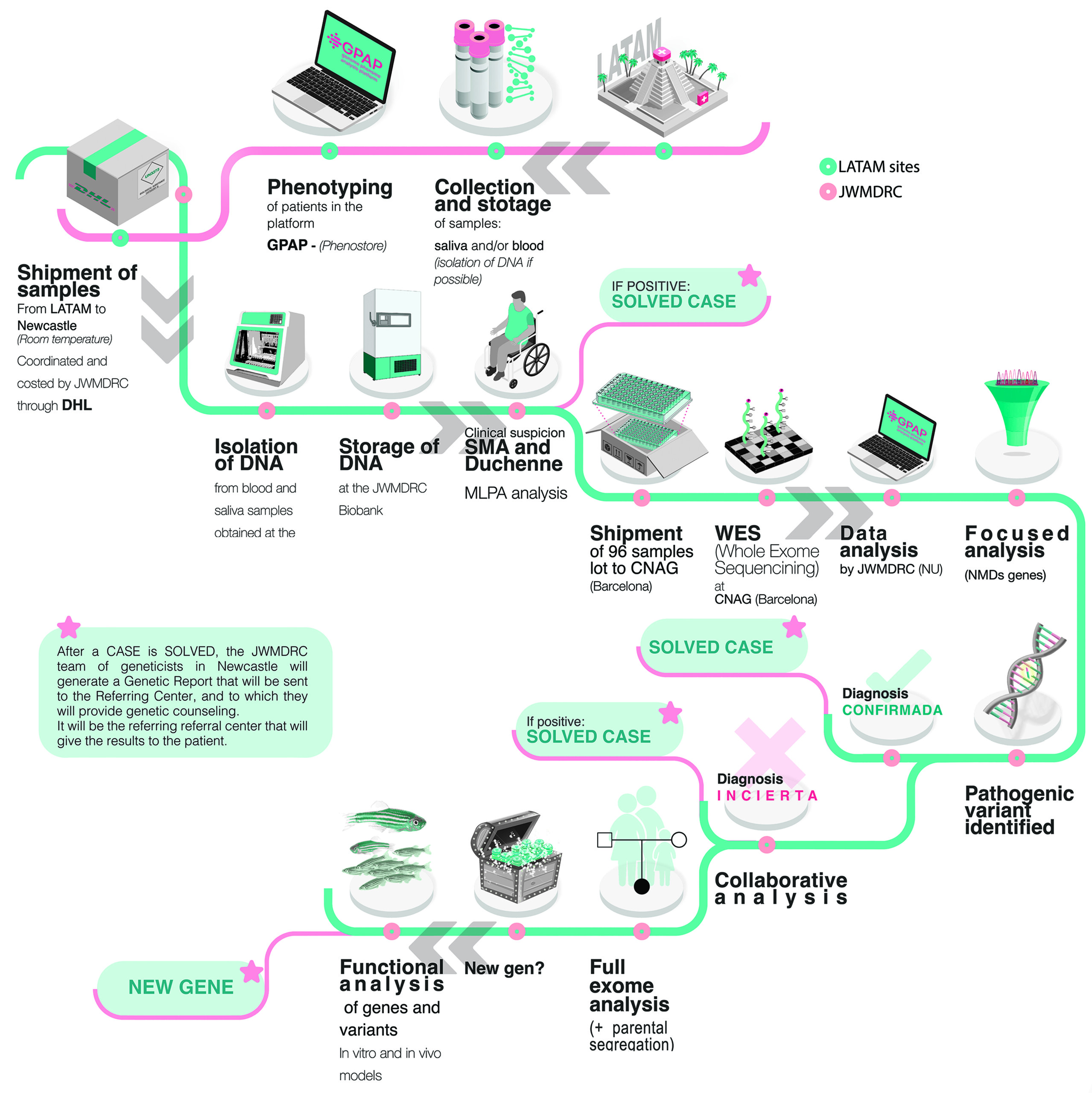
Healthcare professionals at each participating centre will select their patients and ensure that participants sign the informed consent form. They will also be responsible for collecting biological samples (DNA, blood, saliva, or dried blood spots depending on availability). These samples will be sent to and stored at the “JWMDRC Biobank” in Newcastle until analysis. Additionally, they will need to provide demographic and clinical data for each participant, including physical examination findings, results of complementary studies such as blood tests, electrophysiological studies, muscle MRI, and muscle biopsy if available. This data will be entered into an online platform (PhenoStore) that uses the Human Phenotype Ontology (HPO) to standardise clinical information. Whenever possible, samples from parents and other family members, both affected and healthy, will also be collected. The coordination team in Newcastle will manage and cover the costs of shipping the samples from the referring centres to Newcastle.
ANALYSIS STRATEGY:
- Initial Screening:
Depending on the clinical suspicion indicated by the referring physician, the samples will undergo a series of genetic analyses prior to exome sequencing. These include MLPA for Duchenne/Becker muscular dystrophy, Spinal Muscular Atrophy (SMA), and CMT1A neuropathies, as well as recurrent point mutation analysis for those suspected of congenital myasthenia and mitochondrial disease. It is estimated that this analysis will diagnose 20-30% of the cases analysed. The remaining cases will undergo exome sequencing. - Proband-Only Exome Sequencing:
DNA samples from the index cases and their affected relatives will be sent to the National Centre for Genomic Analysis (CNAG) in Barcelona, Spain, where whole exome sequencing and initial bioinformatics processing will be performed. The processed data will be uploaded to the Genome-Phenome Analysis Platform (GPAP) of RD-Connect, where variant filtering will follow standard strategies for rare diseases (e.g., population frequency and variant effect predictors) to focus on those with the highest clinical relevance. Identified variants will be classified according to the ACMG (American College of Medical Genetics and Genomics) guidelines. Data analysis will be conducted in stages. In the first stage, Whole Exome Sequencing (WES) of the index cases and affected relatives will be performed, using an in-silico panel covering more than 600 genes associated with neuromuscular diseases (https://pubmed.ncbi.nlm.nih.gov/36697115/) This initial approach is expected to resolve between 40% and 60% of cases. - Thorough Research:
In a later stage, unresolved cases will be examined more thoroughly, guided by the phenotype and clinical history of each patient. Genes outside the initial panel will be analysed, and/or parents will undergo trio exome sequencing. This trio or family analysis will help clarify complex diagnoses and may identify new genes not yet associated with neuromuscular diseases. - Case Discussions:
After the first round of analysis, case discussions will take place between local collaborators and the coordinating group to evaluate the clinical relevance of the identified variants and plan the appropriate management for each patient. - Data Storage and Access:
All genomic data will be stored on the restricted-access GPAP platform, facilitating collaborative analysis and critical review of clinical and genetic findings.
PUBLICATIONS:
The Latin-SEQ project values the contribution and effort of each and every referring and collaborating centre. To ensure fair and collaborative representation, any general publication by the coordinating team in Newcastle will include each member of the Latin-SEQ Consortium who has contributed samples to the study as a co-author. This inclusion ensures proper recognition of all participants and promotes the visibility of the collective work within the scientific community. Additionally, each centre will have full freedom to lead works and/or presentations of local and individual cases. This publication policy fosters transparency, international collaboration, and scientific advancement, allowing the findings and knowledge generated through the Latin-SEQ project to be widely shared, benefiting the medical and scientific community and patients with neuromuscular diseases.
WHAT IS LATIN-SEQ:
Latin-SEQ is a research project with the primary aim of providing genetic diagnosis to individuals affected by hereditary neuromuscular diseases (NMD) residing in Latin American (LATAM) countries. To achieve this, we utilise a range of genetic techniques, primarily «Next Generation Sequencing» (NGS), but also traditional sequencing using Sanger and MLPA techniques. This is an international and multicentre study involving over 60 hospitals across 18 countries, coordinated by the John Walton Muscular Dystrophy Research Centre (JWMDRC) at Newcastle University, United Kingdom.
Our commitment is to accelerate the genetic diagnosis process for these patients and expand knowledge about the causes and prevalence of neuromuscular diseases in different LATAM populations. Obtaining an early diagnosis offers patients numerous benefits: access to a follow-up plan and personalised therapy, clear expectations about the progression of their disease, the possibility of receiving genetic counselling for family members, the opportunity to be included in patient registries or associations, and participation in clinical trials if available.
Latin-SEQ promotes the creation of a robust network of hospital units and clinical centres with expertise in neuromuscular diseases, catering to both adult and paediatric patients across LATAM.
Latin-SEQ also includes virtual spaces for specialised training, joint discussion of clinical cases, and collaboration on research projects, bringing together neurologists, paediatric neurologists, neurophysiologists, geneticists, physiotherapists, and other healthcare professionals involved in the diagnosis and follow-up of patients with neuromuscular diseases.
OBJETIVES:
WHAT IS EXOME SEQUENCING:
Sequencing is a molecular technique that allows us to analyse a person’s DNA to “read” their genetic code. Previously, methods such as Sanger sequencing were used, which, although accurate, were slower as they only allowed for the sequencing of a single region of DNA (gene) at a time.
Nowadays, we use more advanced techniques like Next-Generation Sequencing (NGS). This new technology is faster and allows for the simultaneous sequencing of large regions of DNA in a short period of time, facilitating and accelerating genetic diagnosis. In the Latin-SEQ project, we use whole exome sequencing to obtain information from all coding regions, which are those that contain the instructions to produce all proteins. By comparing the exomes of an affected individual with a reference sequence, we can identify genetic variants that may be the cause of these neuromuscular diseases. Thanks to this technology, we can offer a faster and more accurate diagnosis, thus improving the care and quality of life of patients.
INCLUSION AND EXCLUSION CRITERIA:
All patients with a suspected genetic neuromuscular disease, even without a diagnosis, can be included in the Latin-SEQ project. There are no restrictions based on sex or age (both adult and paediatric patients are eligible), nor on the type of neuromuscular disease they present (affecting muscle, neuromuscular junction, or nerve). Only acquired diseases or those that cannot be diagnosed through exome analysis (such as DM1/2 and FSHD) will be excluded. The study will prioritise the analysis of families with multiple affected members and patients from ethnic minorities.
RECRUITMENT, SAMPLE COLLECTION, AND PHENOTYPING:
Healthcare professionals at each participating centre will select their patients and ensure that participants sign the informed consent form. They will also be responsible for collecting biological samples (DNA, blood, saliva, or dried blood spots depending on availability). These samples will be sent to and stored at the “JWMDRC Biobank” in Newcastle until analysis. Additionally, they will need to provide demographic and clinical data for each participant, including physical examination findings, results of complementary studies such as blood tests, electrophysiological studies, muscle MRI, and muscle biopsy if available. This data will be entered into an online platform (PhenoStore) that uses the Human Phenotype Ontology (HPO) to standardise clinical information. Whenever possible, samples from parents and other family members, both affected and healthy, will also be collected. The coordination team in Newcastle will manage and cover the costs of shipping the samples from the referring centres to Newcastle.
ANALYSIS STRATEGY:
- Initial Screening:
Depending on the clinical suspicion indicated by the referring physician, the samples will undergo a series of genetic analyses prior to exome sequencing. These include MLPA for Duchenne/Becker muscular dystrophy, Spinal Muscular Atrophy (SMA), and CMT1A neuropathies, as well as recurrent point mutation analysis for those suspected of congenital myasthenia and mitochondrial disease. It is estimated that this analysis will diagnose 20-30% of the cases analysed. The remaining cases will undergo exome sequencing. - Proband-Only Exome Sequencing:
DNA samples from the index cases and their affected relatives will be sent to the National Centre for Genomic Analysis (CNAG) in Barcelona, Spain, where whole exome sequencing and initial bioinformatics processing will be performed. The processed data will be uploaded to the Genome-Phenome Analysis Platform (GPAP) of RD-Connect, where variant filtering will follow standard strategies for rare diseases (e.g., population frequency and variant effect predictors) to focus on those with the highest clinical relevance. Identified variants will be classified according to the ACMG (American College of Medical Genetics and Genomics) guidelines. Data analysis will be conducted in stages. In the first stage, Whole Exome Sequencing (WES) of the index cases and affected relatives will be performed, using an in-silico panel covering more than 600 genes associated with neuromuscular diseases (https://pubmed.ncbi.nlm.nih.gov/36697115/) This initial approach is expected to resolve between 40% and 60% of cases. - Thorough Research:
In a later stage, unresolved cases will be examined more thoroughly, guided by the phenotype and clinical history of each patient. Genes outside the initial panel will be analysed, and/or parents will undergo trio exome sequencing. This trio or family analysis will help clarify complex diagnoses and may identify new genes not yet associated with neuromuscular diseases. - Case Discussions:
After the first round of analysis, case discussions will take place between local collaborators and the coordinating group to evaluate the clinical relevance of the identified variants and plan the appropriate management for each patient. - Data Storage and Access:
All genomic data will be stored on the restricted-access GPAP platform, facilitating collaborative analysis and critical review of clinical and genetic findings.
PUBLICATIONS:
The Latin-SEQ project values the contribution and effort of each and every referring and collaborating centre. To ensure fair and collaborative representation, any general publication by the coordinating team in Newcastle will include each member of the Latin-SEQ Consortium who has contributed samples to the study as a co-author. This inclusion ensures proper recognition of all participants and promotes the visibility of the collective work within the scientific community. Additionally, each centre will have full freedom to lead works and/or presentations of local and individual cases. This publication policy fosters transparency, international collaboration, and scientific advancement, allowing the findings and knowledge generated through the Latin-SEQ project to be widely shared, benefiting the medical and scientific community and patients with neuromuscular diseases.